Lessons from our approach to bioprospecting in ticks
Contributors (A-Z)
Lessons from our approach to bioprospecting in ticks
Purpose
Our first and longest-running startup incubation project at Arcadia is Trove. At Trove, we set out with an ambitious goal of translating insights from venomous, blood-feeding ticks into therapeutics. Ticks can feed for several days at a time, which means they’ve evolved ways to suppress host responses like itch. We bet on this biology by mining for interesting tick molecules that we hoped could source some much-needed new anti-itch solutions.
After a nearly five-year effort, we’re winding Trove down because we don’t see a reasonable path forward. Certainly not on a timeline that made sense for us, given the value we expect to realize from the venture should increase with its investment cost. This effort was a particularly hard one to ice given everything we put into it, and everything we overcame as a team. However, we feel good about our decision. We left no (reasonable) stone unturned, and we learned a ton.
We encountered not only expected technical difficulties, but also roadblocks exacerbated by our starting assumptions and lack of constraints. We’ve summarized our top lessons in this pub. Some of the principles are fairly obvious, but they’re still worth repeating, especially for other scientists who are considering an entrepreneurial venture for the first time.
Share your thoughts!
Feel free to provide feedback by commenting in the box at the bottom of this page or by posting about this work on social media. Please make all feedback public so other readers can benefit from the discussion.
We’ve put this effort on ice! 🧊
#DeadEnd #HardToScale #MissingExpertise
This was a tough project! We still believe that ticks represent an evolutionary cache of potential therapeutics. But while looking for those molecules, we encountered technical hurdles, strategy challenges, and gaps in our expertise that ultimately convinced us to ice the project.
Learn more about the Icebox and the different reasons we ice projects.
Hunting for anti-itch therapeutics in ticks
Motivation
Itch is a symptom associated with a wide range of clinical indications and can sometimes be so severe that it is debilitating. While some therapeutics can ameliorate itch symptoms, there remains significant unmet need in patients experiencing chronic itch, even with newly approved therapeutics on the market. Part of the challenge is that we don’t yet know the full mechanistic basis of itch and haven’t yet identified the key causal determinants [1][2]. In addition, there are actually multiple mechanisms of itch [3], which stem from both overlapping and distinct pathways. Bottom-up approaches based on our molecular and cellular understanding haven’t been successful so far.
We decided to tackle this problem from the other side. Namely, instead of trying to dissect the mechanisms of itch, we looked to biology for pre-tested solutions as part of a startup incubation project called Trove. Our hypothesis was this: ticks bite humans and evade detection by our skin defenses, so they might harbor therapeutically useful molecules that reduce itch.
Our strategy
We chose Amblyomma americanum ticks because they’re known to bite humans, are readily available to purchase through several sources, and are on the larger side and therefore advantageous for generating lots of starting material. We built an experimental pipeline that involved extracting material from A. americanum, chromatographically separating that material into pools of molecules (fractions), empirically testing different fractions for anti-itch activity in an in vivo mouse scratch assay, and analyzing fractions with apparent anti-itch activity to identify their component molecules (Figure 1). We reasoned that starting with a behavioral phenotype that most closely resembles the experience of human itch would help us more effectively filter for molecules with the activity we cared about.
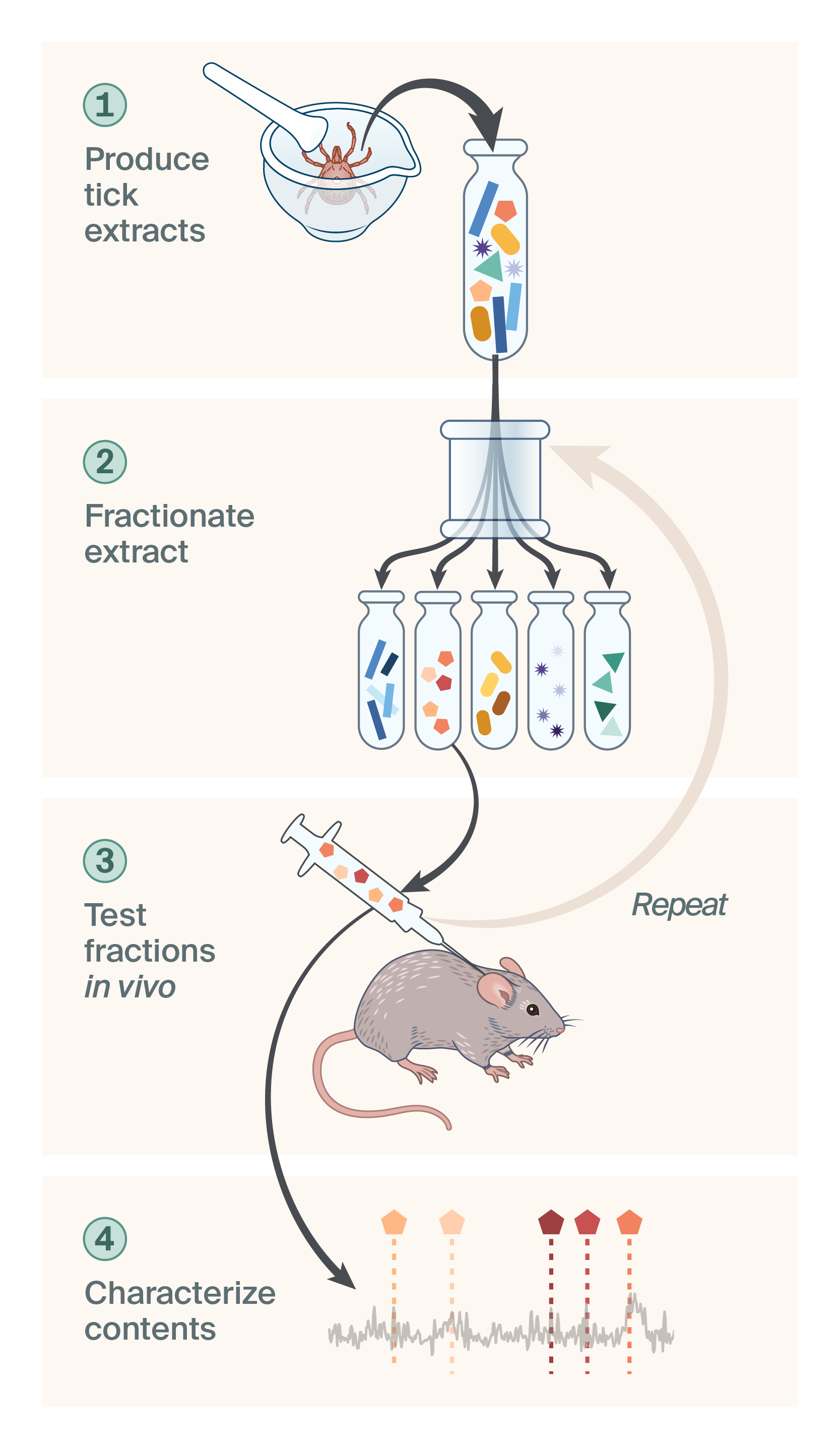
Trove’s discovery pipeline.
We designed a discovery pipeline that involved 1) extracting material from ticks, 2) separating that material into pools of molecules (fractions), 3) testing fractions for anti-itch activity in vivo, and 4) characterizing the molecules in active fractions.
Here’s a little more detail about our approach:
- Producing tick extracts
To find bioactive molecules, we initially dissected salivary glands from ticks and prepared extracts from the glands. Salivary glands store many of the molecules that are released into the host during feeding, so we hoped to enrich for anti-itch activity by starting here. We had some encouraging early results from these extracts, but dissection was laborious and quickly became a major bottleneck. To move faster and generate more starting material, we experimented with preparing aqueous and organic extracts from frozen whole ticks. In vivo tests indicated the aqueous extract was strongly antigenic (perhaps not surprising), which led to noisy readouts, but the organic extract was not. We moved forward with testing and fractionating the organic extract. - Fractionating the extract
To find active molecules in the organic extract, our strategy was to chromatographically separate the material into different pools of molecules (fractions) using HPLC. Both C18 semi-preparative and preparative columns were successful at separating lipophilic molecules across the spectrum. Then we combinatorially tested the fractions in our in vivo activity assay. - Testing fractions in an in vivo itch assay
To look for anti-itch activity in the fractions, we developed a mouse behavioral assay to measure responses to acute itch stimuli. We believed that evaluating itch at the whole-organism level would capture the full complexity of the neuroimmune network. This was particularly critical for dissecting itch, which is mediated by an interplay between many cell types in the immune system, the skin barrier, and the peripheral and central nervous systems. Using an in vivo assay allowed us to be more mechanism-agnostic, which we thought would improve translatability. - Characterize chemical composition of promising fractions
After identifying active fractions, we sub-fractionated those molecules and repeated the testing process. Our plan was to repeat this cycle of testing and sub-fractionating until we reduced the complexity of the fractions sufficiently to use mass spectrometry to begin characterizing the molecules. To complement this approach, we also explored Raman spectroscopy as a way to track the spectroscopic fingerprint of different classes of molecules through a series of fractions.
How far did we get?
Even though we’re icing this project, we’re happy to report that we do think 1) ticks harbor anti-itch activity and 2) it’s possible to find this activity empirically. We ultimately discovered organic lipid fractions that quantitatively reduced scratching in mice. By refining the in vivo mouse assay and applying an automated workflow [4] to speed it up, we were able to sort through an estimated 20,000 unique small molecules in the organic fraction to < 100 putative therapeutic candidates that needed to be investigated.
However, to get to the next stage of discovery, in which we could experimentally test our candidates, required a level of chemistry expertise and throughput that we don’t currently have. While the personnel gap is theoretically solvable, we believe this project needs significant chemistry expertise and resources to develop innovative, cutting-edge solutions for natural product discovery. It likely makes more sense to build this infrastructure at a company pursuing multiple lines of anti-itch therapeutic discovery at once. If we were to begin orthogonal avenues of discovery, we’d need new assays, because our current experimental throughput is too low. We did some exploration into what such orthogonal assays might look like [5][6], but ultimately don’t feel that we have enough scientific understanding about the biology of itch to design creative and effective measures of anti-itch activity.
High-level takeaways
Frontload the right risks
Many of our pivotal realizations came too late, and we underestimated which risks were the most important and disqualifying. We got some of them right, but missed a few that were key.
Importantly, experimental robustness is obviously critical for any science we do, but we didn’t fully appreciate how difficult this might be for itch biology. A lot of drug development in itch tends to start with molecular or cellular phenotypes that never translate into real effects on itch-induced scratching in animals. We had a stringent behavioral assay for itch in mice that told us we were really on to something, but such scratch assays are expensive, noisy, and slow. Together, these qualities became deal-breakers, given how many iterations are required for fractionation-based natural product discovery.
We should have prioritized assessing whether we could see convergence across multiple orthogonal assays with a range of throughput and cost. We started this process a bit late and never succeeded.
Why not? There’s still too much we don’t yet understand about the precise mechanistic basis of itch to confidently connect molecular and cellular assays with behavioral ones, which became a key challenge to our discovery pipeline. Itch can arise from a number of mechanistically distinct pathways, making conclusions messy and prone to misinterpretation. Plus there are known differences in both the neuro- and immune-biology of mice versus humans, further exacerbating this challenge.
In retrospect, we should have created a checklist of all the features required for experimental rigor before moving forward. If we were to try again, this would be our list:
- ≥ 1 assay outside of behavioral tracking, to allow for orthogonal testing
- ≥ 1 assay that is high-throughput and accommodates small amounts of material
- ≥ 1 assay that is high-throughput and agnostic to specific itch mechanism or a small set of assays that we know span the full spectrum of itch mechanisms of interest
- Confirmation that orthogonal assays report on same behavioral phenotype (scratching)
Constraints are your friend
One of the defining challenges came from our attempt to stay flexible on a number of fronts, some of which stems from our basic science training. We thought, if you don’t know what particular endstate could be possible, you should just keep all your options open. This was the wrong idea for a therapeutics venture. Some outcomes are worse than others, and a subset of those may even be non-starters.
A major example is our openness to what type of molecular modality we might find in ticks for blocking itch. We made sure to mine for options across all tick fractions that spanned biologics and small molecules. In reality, these two classes of hits have very different and important implications for what comes next. We wanted to keep exploring both in the lab. Operationally speaking, we did have to go ahead and make a bet on the kind of infrastructure and talent profiles required. In keeping with the broader biology-focused strategy of Arcadia’s platform, we biased towards biologics.
When our work pointed us toward lipid fractions, we underestimated how much more complex and resource-intensive this non-biologics direction would be. Small molecules don’t have a straightforward reference map like proteins have with genomes or transcriptomes. Unless you find something with a characterized chemical neighbor, there are limited paths to de novo characterization the hard way (natural product isolation, mass spectrometry, nuclear magnetic resonance). We found ourselves staring up a mountain that we didn’t have the gear to climb.
We now see that we really had two options: make an upfront modality commitment so we were operationally prepared to pounce if we found something exciting. Or design for optionality, which would have required a far more aggressive upfront investment in team-building and capabilities to cover multiple scenarios. We did neither.
Always test the scientific assumptions yourself
A critical blind spot was our reliance on academic findings that turned out to be non-reproducible in our hands. We knew this would be a challenge, but we didn’t fully appreciate its enormity (and cost).
We should have made a list of all the foundational scientific claims and assays we were counting on and built a programmatic effort early on to make certain we could lean on them. This should have been a central and front-loaded pillar of our work. Instead, we ended up with layers of uncertainty at each turn: uncertain assays, uncertain readouts, uncertain conclusions. And as we began talking about our findings to more scientists, more individuals revealed related controversies and disagreements within their fields that were excluded from published articles. Below are the key issues that slowed us down — we encourage others in the field to watch out for these potential stumbling blocks:
- Published scratch behavior for a given itch-inducing molecule wasn't always reproducible in our hands
- For some itch inducers, variance in scratching was higher than expected (both within and between individuals). For example, we found that some animals barely responded to serotonin injections, while others scratched aggressively
- Publications often failed to report important experimental details, like the volume of injections or the age/sex of the mice
As scientists, we need to do a better job of more explicitly bringing out uncertainties and debate, and to craft methods sections that scientists outside of a particular field can reproduce. Partial understanding turned out to be worse than none at all. Building something that works is a different bar than publishing an article. Assume the house is on sand unless you’ve poured the foundation yourself.
Natural product discovery is really hard
In theory, natural product discovery has a ton of potential, given how vast a solution space biology has explored, both functionally and chemically. Many of the most successful therapeutics were either directly or indirectly inspired by natural molecules. But practically speaking, it’s very risky. Sometimes, hard things are just super hard.
For example, without data-rich pipelines or predictive models, fractionation-based hacking is expensive, slow, and dependent on luck. In our case, this was further amplified by the fact that we were largely depending on a mouse behavioral assay.
It’s also very difficult to finish that final leg of molecular isolation. The standard empirical process-of-elimination approach can reliably reduce your sample complexity from tens of thousands to hundreds or even dozens of molecules. In our case, crossing the final divide to find that one winning hit proved insurmountable. It was difficult to get enough material to overcome noisy, inconsistent readouts. Because we didn’t have enough clues to predict our chemical structure, we'd need orders of magnitude more material to conduct characterization studies with NMR.
The field needs new ways of making natural product discovery more deterministic, or at least navigable. We made some headway on this front by pairing fractionation with Raman spectroscopy, a tracking approach that was cheaper and faster than mass spectrometry. While this wasn’t quite enough to overcome our obstacles, we’re hoping new chemical discovery technologies developed elsewhere could revive this effort.
Even if we succeeded in isolating and characterizing the structure of our small molecule, we faced a long and uncertain road ahead in synthesizing and manufacturing it at scale for development. Combining that reality with our multitude of challenges and unknowns, we simply didn’t believe that we could reasonably expect to get this to the finish line soon enough.
AI writing tool usage
We used ChatGPT to suggest wording ideas (we then chose which small phrases or sentence structure ideas to use), to expand on summary text that we provided (we then edited the text it produced), and to help clarify and streamline text that we wrote. We also used Notion AI to suggest wording ideas and then chose which small phrases or sentence structure ideas to use.
What’s next?
We’re still believers in the core idea: that nature’s molecular diversity holds untapped therapeutic potential, especially when informed by ecological interactions between humans and the natural world. Specifically, we think that ticks and other venomous creatures have a lot to offer that others should explore. We’ve shared several of our tools and resources from Trove’s work in the hopes that others could either do more basic research on the topic or perhaps try their hand at a startup venture if they have solutions to our roadblocks. We'd love for others to make use of:
We’re also wrapping up a research pilot exploring the use of Raman spectroscopy as a way to quickly and cheaply track sample composition during fractionation. We believe this could speed up any tracking work that is typically dependent on MS. Stay tuned for a pub on this soon!
- Acknowledgments
- It took a village to build Trove. Thank you to everyone who helped us over the years! We’d like to give a special shoutout to the following people for going above and beyond: Claire Kwon, Tori Doran, Peter Thuy-Boun, Althea Stillman, Alba Peinado, Feridun Mert Celebi, Lara Ortiz-Luis, Allan Basbaum, Thea Mauro, Kelly Moynihan, Becky Pferdehirt, Xintong Dong, Joao Braz, Juan Salvatierra, Sakeen Kashem, Mahsa Sadeghi, Hans Hofland, David Thomson, Bill Fenical, Ju-eun Jeon, and Lisa Coburn.
And last, but not least, all the Arcadians past and present who jumped in to help whenever we needed. Even for manual mouse counts over Christmas!
- It took a village to build Trove. Thank you to everyone who helped us over the years! We’d like to give a special shoutout to the following people for going above and beyond: Claire Kwon, Tori Doran, Peter Thuy-Boun, Althea Stillman, Alba Peinado, Feridun Mert Celebi, Lara Ortiz-Luis, Allan Basbaum, Thea Mauro, Kelly Moynihan, Becky Pferdehirt, Xintong Dong, Joao Braz, Juan Salvatierra, Sakeen Kashem, Mahsa Sadeghi, Hans Hofland, David Thomson, Bill Fenical, Ju-eun Jeon, and Lisa Coburn.
References
Share your thoughts!
Feel free to provide feedback by commenting in the box at the bottom of this page or by posting about this work on social media. Please make all feedback public so other readers can benefit from the discussion.