Modeling human monogenic diseases using the tunicate Ciona intestinalis
Contributors (A-Z)
Modeling human monogenic diseases using the tunicate Ciona intestinalis
Purpose
We developed a framework for selecting disease–gene pairs from our organism selection dataset for experimental testing [1]. Here, we apply that framework to genes in the tunicate Ciona intestinalis (recently disambiguated into two species, Ciona intestinalis and Ciona robusta [2]) to identify three genes of interest. We briefly introduce the life history and available genetic tools in Ciona. We also provide examples that explain our rationale for rejecting or pursuing individual genes.
Researchers developing genetic models of human disease using animals, particularly tunicates, may find it informative to read about how we considered experimental design trade-offs. In addition, we develop visualization tools for single-cell and bulk RNA-seq datasets for Ciona that others may find useful when designing experiments.
- All code related to this pub is available in this GitHub repo. Code specific to C. intestinalis is in these analysis notebooks and this set of visualization scripts.
Share your thoughts!
Feel free to provide feedback by commenting in the box at the bottom of this page or by posting about this work on social media. Please make all feedback public so other readers can benefit from the discussion.
About the organism
Ciona intestinalis is a tunicate (sea squirt) and a well-established model organism in developmental biology [3]. Tunicates are marine invertebrate chordates that represent the closest living relatives to vertebrates [4], making them invaluable for understanding chordate evolution and development [5]. This solitary ascidian is studied extensively to understand fundamental mechanisms of embryonic development, cell fate specification, and the evolutionary origins of vertebrate characteristics.
Adult Ciona are sessile filter-feeders with a distinctive sac-like body plan, but their free-swimming tadpole larvae possess classic chordate features, including a notochord, dorsal neural tube, and muscular tail [6] (Figure 1, A). The larvae undergo rapid development, completing embryogenesis in just 18 hours at 18 °C [7], making them ideal for developmental studies. Following a brief planktonic phase, tadpole larvae settle and undergo dramatic metamorphosis, resorbing their tail and notochord while developing into the sessile adult form. This life cycle transformation provides unique insights into chordate body plan evolution and the developmental programs underlying major morphological transitions.
The Ciona embryo is particularly tractable for cell lineage studies due to its invariant cleavage pattern and relatively small cell number [8][9]. Each blastomere's fate can be precisely traced, and the embryo's optical transparency allows for detailed observation of cellular behaviors during development.

Overview of diligence on Ciona intestinalis.
(A) Illustration of life cycle of Ciona and schematic of available genetic tools and types of human alleles that we expect to be able to model, given available tools. Sessile adults spawn embryos, which develop into free-swimming tadpole larvae with chordate-like features. These tadpoles settle and metamorphose into the adult form.
(B) Illustration of manual diligence outcomes listing the number of genes eliminated by each failure mode. The initial shortlist was nine genes, three of which were selected for experimental testing.
State of technology
Ciona has robust genetic tools and genomic resources.
- Genetic tools. Ciona has well-developed genetic manipulation capabilities. Electroporation-mediated transgenesis is routine, allowing for tissue-specific gene expression studies using established promoter elements [10][11]. CRISPR-Cas9 genome editing is highly efficient in this system, enabling targeted mutagenesis [12][13]. However, some barriers to reproducible CRISPR knockouts in this organism include the high degree of genome heterozygosity (1.1–1.2%) [14] and the fact that many laboratories obtain Ciona embryos through environmentally collected adult samples, rather than working on stable lines with a closed life cycle.
Previous reports have indicated that it’s possible to generate CRISPR knock-ins within Ciona embryos [15], but the experts we consulted with haven’t been able to reproduce this work in their lab. As such, we focused on CRISPR knockouts as our main tool for generating genetic models.
Generating mutant lines in Ciona is possible with sufficient time and resources. However, the experts we consulted with aren’t well equipped to close the life cycle on knockouts, which requires metamorphosis, rearing, and maintenance of adult animals, so we’ve decided to focus on phenotypes in G0 knockout embryos.
- Phenotypic assays. Ciona embryos and larvae are highly amenable to experimental manipulation and observation (for details about genetic tools and the size of the organism, see [16]). Thousands of ~0.1mm embryos can be collected and electroporated at once, allowing for high-throughput experiments [11]. Standard protocols exist for in situ hybridization [17], immunohistochemistry [18], and generation of reporters using transposon-mediated transgenesis [19]. The transparent embryos are particularly well-suited for live imaging approaches.
A unique feature of Ciona development is that knockouts of normally essential genes don’t usually lead to embryonic death, due to a lack of apoptotic processes in the embryo. This means that some genes that would be lethal in human cells might have a recoverable phenotype in Ciona.
- Gene expression. Comprehensive transcriptomic resources are available for C. intestinalis, including developmental time-course RNA-seq data covering embryonic and larval stages [20]. Single-cell RNA sequencing has provided detailed cell-type-specific expression profiles throughout development, revealing the molecular basis of cell fate specification and differentiation programs in this chordate model system.
Diligence results
We manually diligenced 27 different genes in Ciona intestinalis to arrive at nine potentially actionable genes for pilot experiments. A summary of the diligence results can be seen in Figure 1, B.
You can see a copy of the working document we used to catalog our thoughts for each Ciona gene we diligenced here.
Example genes we rejected
In this section, we explain why we rejected a few genes from C. intestinalis to show how we applied our selection criteria.
CACNB2 (calcium voltage-gated channel auxiliary subunit beta 2)
Model not feasible
This subunit of a voltage-gated calcium channel is implicated in a subtype of Brugada syndrome, a disease that causes death by sudden heart failure and is detectable by abnormalities in electrocardiogram signals [21]. All cases of this disease recorded in studies in OMIM were heterozygous missense mutations. Because we can't easily generate targeted mutations in Ciona, we decided not to pursue this gene.
RP9 (RP9 pre-mRNA splicing factor)
Disease association concerns
This protein is implicated in a subtype of retinitis pigmentosa (RP), a disease that causes gradual loss of eyesight due to the death of retinal cells. While a particular variant (His137Leu) has been reported in the literature as causing RP [22], follow-up work seems to indicate that sequencing results identifying this variant may actually come from a different pseudogene locus within the genome, which could have been amplified in previous assays [23][24]. Because there's a lack of confidence in identifying variations in this gene as causative of RP, we decided not to pursue this gene.
MLYCD (Malonyl-CoA decarboxylase)
Lack of model advantage
This protein is involved in fatty acid metabolism, where it breaks down malonyl-CoA into acetyl-CoA and carbon dioxide [25]. Loss of this protein leads to metabolic defects across organ systems. When considering how Ciona might model this disease, we also considered whether identifying molecules that help clear malonyl-CoA in human in vitro cell culture systems might be more efficient. It’s possible that modeling in Ciona might uncover phenotypes that wouldn’t be obvious in cell lines or less appropriate models. However, the experts we consulted also previously indicated that metabolic phenotypes or others involved in growth may not be well-modeled by the fast-developing embryos of Ciona. For these reasons, we decided not to pursue this gene.
Our initial shortlist
Here, we describe a few of the best-supported genes to model in Ciona.

Structural comparisons of top-selected shortlist proteins to human proteins.
(A) FCH and mu domain-containing endocytic adaptor 1 [flexible alignment generated using jFATCAT for visualization purposes (see the “Methods” section of our previous pub for more details [26])]
(B) Phosphoglucomutase 3
(C) NCK-associated protein 1-like protein
(D) NAD(P)HX epimerase
Tan proteins are predicted human structures; gold proteins are predicted C. intestinalis homolog structures. “Trait distance” is a multivariate Mahalanobis distance between pairs of proteins calculated based on 10 physicochemical properties (see the “Methods” section of our previous pub for more details [26]). “Portfolio rank” is the relative rank of this protein compared to all other proteins found in the organism selection/the Zoogle portal. TM-score is a standard measure of solid-body protein structural similarity [27]. In general, TM-scores of 0.5 or above indicate the same fold.
FCHO1 (FCH and mu domain-containing endocytic adaptor 1)
This protein is a nucleator of clathrin-coated pits [28]. Loss of function through missense, nonsense, or splice mutations leads to an immunodeficiency characterized by recurrent infections due to T- and B-cell dysfunction and usually death in childhood [29][30].
The Ciona match from our organism selection pipeline shows both FCHO1 and FCHO2 as strong hits from Foldseek, but our interpretation of this similarity is confounded because this family of proteins is large and appears to contain many disordered regions, which makes it unsuitable for comparison using rigid-body structural comparison approaches such as the TM-score (Figure 2). The Ciona homolog of FCHO1 appears to be expressed throughout the embryo during development (Figure 3).
Previous work in Ciona has investigated the function of clathrin endocytosis in the morphogenesis of the notochord [31]. During this process, a lumen is formed between adjacent cells in the notochord. Drugs that target clathrin-mediated endocytosis, including chlorpromazine, pitstop 2, and Dynasore, disrupt the formation of this lumen [31]. Given the strong possibility of a similar phenotype when mutating Fcho1 in Ciona, we decided that this gene would be tractable for downstream analyses.
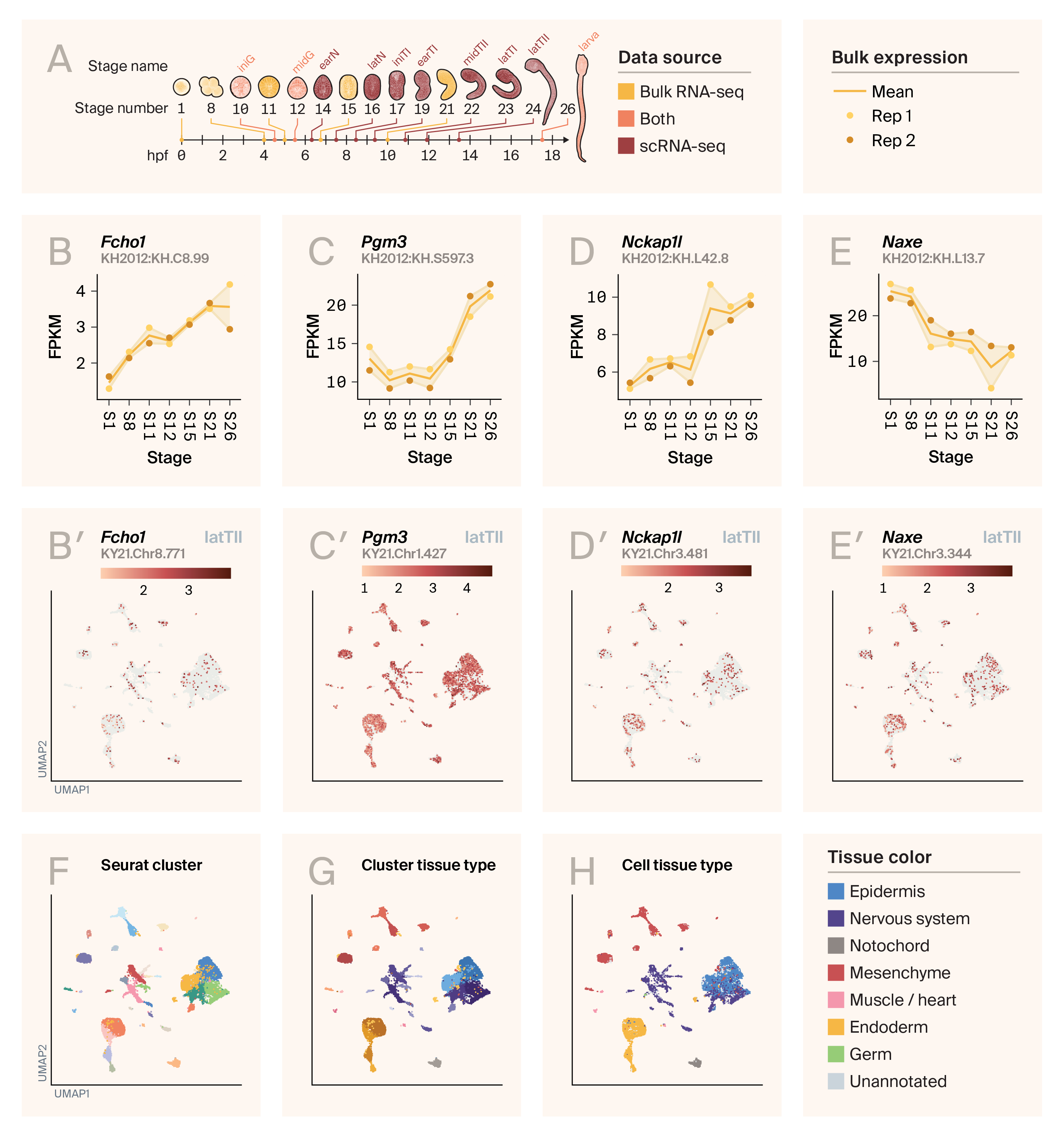
Bulk and single-cell RNA-seq expression in Ciona intestinalis embryogenesis.
(A) Illustration of Ciona’s developmental stages. Yellow stages are found in the bulk RNA-seq dataset from ANISEED. Red stages are found in the single-cell RNA-seq data from Cao et al. [20] and the Piekarz reanalysis. Stages colored in orange are shared between the datasets.
hpf: hours post-fertilization, FPKM: fragments per kilobase-million, latTII: late tailbud II, UMAP: uniform manifold approximation and projection
(B–E) Expression of Fcho1 (B), Pgm3 (C), Nckap1l (D), and Naxe (E) in the ANISEED bulk RNA-seq dataset. ANISEED data uses the KH2012 identifier, which is displayed in each panel.
(B′–E′) Expression of Fcho1 (B′), Pgm3 (C′), Nckap1l (D′), and Naxe (E′) in the Cao / Piekarz datasets, for all cells in stage LatTII. The Piekarz dataset uses the KY2021 identifier, which is displayed in each panel.
(F) Cells from panels B′–E′, colored by Seurat cluster identified in the Piekarz reanalysis.
(G) Cells from panels B′–E′, colored by Seurat cluster identified in the Piekarz reanalysis based on hues that represent each primary tissue type found in the dataset.
(H) Cells from panels B′–E′, colored by the individual tissue type label for each individual cell, from the Cao et al dataset.
For bulk expression of additional genes, see this supplementary figure.
PGM3 (phosphoglycomutase 3)
Phosphoglycomutase is a crucial enzyme involved in glycosylation processes that converts GlcNAc-6-phosphate to GlcNAc-1-phosphate [32]. Heterozygous missense, frameshift, and nonsense mutations result in immunodeficiency characterized by recurrent infections and morphological defects across organ systems [33][34][35]. Mice with partial loss of function show defects in hematopoiesis, while homozygous loss of function is embryonic-lethal [32].
PGM3 is the top hit to the Ciona protein in FoldSeek searches and has a very high TM-score of 0.98 (Figure 2). The Ciona homolog is also expressed in a diversity of cell types across development (Figure 2).
Previous work has characterized the degree of glycosylation of proteins in Ciona embryos across developmental stages using mass spectrometry [36]. This work observed many glycosylation patterns that contain the substrates of PGM3, suggesting that its activity would be necessary for the production of these moieties. Disruption of glycosylation through drugs like NGI-1 and tunicamycin also appears to result in defects in notochord lumen formation [37]. Due to the strong evidence for the importance of glycosylation in Ciona development, particularly of the notochord, we were interested in pursuing this gene for experimental testing.
PGM3 is also being investigated as a target for treating breast cancer for its role in the hexosamine biosynthetic pathway (HBP) [38]. A commercially available PGM3 inhibitor, FR054, is available, potentially letting us cross-validate our knockouts with chemical treatments and see how similarly the Ciona homolog responds to this drug.
NCKAP1L (NCK-associated protein 1-like protein)
This protein, also known as HEM1, is a subunit of a larger scaffold that supports the wave regulatory complex (WRC), which itself regulates the actin cytoskeleton [39]. Missense and splice abnormalities in this protein lead to immunodeficiency characterized by recurrent viral and bacterial infections, combined with autoimmunity characterized by excessive inflammation [40][41]. Patient cells appear to have defects in lamellipodia, which are hypothesized to impede proper T-cell development.
The Ciona homolog appears to be a strong structural match to both NCKAP1 and NCKAP1L (TM-score = 0.96) (Figure 2). NCKAP1 is a homolog of NCKAP1L; NCKAP1L is distinct in its high expression in the human immune system. The Ciona homolog appears to be expressed across developmental stages and tissue types (Figure 2).
Lamellipodia are hypothesized to play a role in Ciona notochord lumen development and appear to form at the leading edge of notochord cells during lumen formation [42]. Disruption of the actin cytoskeleton through drugs such as latrunculin B and blebbistatin disrupts notochord lumen morphology, suggesting that proper actin dynamics are required for this process. For these reasons, we were interested in testing whether Nckap1l knockout may have an assayable phenotype in Ciona.
NAXE [NAD(P)HX epimerase]
This metabolic enzyme is necessary for detoxifying metabolic byproducts (S-NADHX, R-NADHX, and cyclic NADHX) of the essential coenzymes NADH and NADPH back into their original, useful states [43]. Missense, nonsense, frameshift, and splice mutations in NAXE lead to a syndrome in which patients, usually infants, experience metabolic crisis after developing a fever due to depleted NADH/NADPH and accumulation of NADHX [44][45][46]. Most otherwise phenotypically normal patients die as a result of this syndrome due to damage to the brain from encephalopathy or leukoencephalopathy.
Curiously, work on mice knockouts of Naxe (also known as Apoa1bp or A1bp due to its additional function in binding apolipoprotein A1) doesn’t appear to show strong effects on lifespan or NADH levels [47]. Mutations of this gene in baker’s yeast (Saccharomyces cerevisiae) [48] and in thale cress (Arabidopsis thaliana) [49] indicate that Naxe isn’t essential for these organisms. Work on this disease could benefit from additional models.
The Ciona homolog of NAXE is a strong structural match to the human homolog (TM-score = 0.79) and is expressed broadly across cell types throughout development. Given its core metabolic function and lack of effective existing models, we were interested in seeing whether we could model NAXE function in Ciona.
Recent work has suggested that treating patients with precursors of NADH, such as nicotinamide and niacin, could help relieve metabolic crisis [45]. Given these data, we were also interested in testing whether these molecules could rescue any potential phenotypic consequences of NAXE knockout in Ciona.
Other hits
Several other genes — RBBP7, GAS2L2, DHODH, CWF19L1, and VWA8 — also made our short list (Table 1).
HGNC gene symbol (UniProt ID) | Human protein name | Associated human disease (OMIM) | Ciona protein UniProt ID | Ciona KH identifier | Ciona KY identifier | Possible Ciona phenotypes |
FCHO1 | FCH and mu domain containing endocytic adaptor 1 | KH.C8.99 | KY21.Chr8.771 | Notochord lumen defects | ||
PGM3 | Phosphoglucomutase 3 | KH.S597.3 | KY21.Chr1.427 | Notochord lumen defects | ||
NCKAP1L | NCK associated protein 1-like protein | KH.L42.8* | KY21.Chr3.481 | Notochord lumen defects | ||
NAXE | NAD(P)HX epimerase | Encephalopathy, progressive, early-onset, with brain edema and/or leukoencephalopathy | KH.L13.7 | KY21.Chr3.344 | Neurodevelopmental defects | |
RBBP7 | RB binding protein 7, chromatin remodeling factor | KH.C8.192 | KY21.Chr8.358 | Gross morphogenetic defects | ||
GAS2L2 | Growth arrest specific 2-like 2 protein | KH.C11.467 | KY21.Chr11.229 | Not evaluated | ||
DHODH | Dihydroorotate dehydrogenase (quinone) | KH.C3.677 | KY21.Chr3.998 | Not evaluated | ||
CWF19L1 | CWF19 like cell cycle control factor 1 | KH.L9.32 | KY21.Chr2.1476 | Not evaluated | ||
VWA8 | Von Willebrand factor A domain-containing 8 | KH.C4.767 | KY21.Chr4.691 | Not evaluated |
Summary of the relevant diseases and our initial guess for phenotypes for each short-list gene.
* = not found in KH-KY-mapping Excel spreadsheet (see Methods).
Expert feedback and our final shortlist
We sought expert feedback on our short list from Alberto Stolfi, a tunicate researcher at the Georgia Institute of Technology, one of the external scientists we consulted as part of this work. After sharing our initial shortlist of genes, our expert advisor noted that many Ciona genes exhibit maternal expression, which can impede the ability to observe phenotypes in CRISPR knockout embryos. Maternal expression in Ciona, as observed in bulk RNA-seq data and according to experts, is generally characterized by high expression at the earliest developmental stages without a substantial increase at later developmental stages. For example, NAXE expression appears to be highest at the one-cell stage and shows a continual decrease in relative expression until hatching (Figure 3). For this reason, we decided not to pursue NAXE for our initial experiments. Other genes we decided not to pursue due to the potential for maternal expression were Gas2l2, Dhodh, and Cwf19l1 (see supplementary figure). While RNAi [50] and morpholino [51] methods are also feasible in Ciona and able to suppress maternal gene expression, we decided to stick to a single method of genetic manipulation for our experimental plan.
Two homologs from our analyses seemed not to have strict maternal expression: Rbbp7 and Vwa8. One function of RBBP7 is as a member of human polycomb repressive complex 2 (PRC2) [52]; morpholinos against the Ciona homolog of Enhancer of zeste (E(z)), another PRC2 component, appear to have morphological consequences throughout development [53]. VWA8 is a mitochondrial matrix-targeted protein with ATPase activity associated with a subtype of retinitis pigmentosa [54]. Morpholino knockdown of zebrafish Vwa8 causes gross morphological defects and degraded retinal cells [54][55]. For these two genes, we weren’t sure exactly what developmental stages, tissues, or cell types would present a clear phenotype. Identifying a phenotype might require first evaluating gene expression through in situ hybridization or transgenic expression, and then generating knockouts and screening for defects across development, which could be time-consuming.
After discussion with experts, we decided that it would be more efficient to focus on a single, well-established assay in Ciona, rather than to survey for potentially unknown phenotypes across development. Broader phenotypic profiling in Ciona could be time-consuming or difficult to develop, given that phenotypes could emerge in various tissues across developmental time. Focusing our work on phenotypes in the notochord also allows us to control for the possibility of non-cell-autonomous effects by generating tissue-specific knockouts. Based on this assay choice, we decided to focus on three genes: Fcho1, Pgm3, and Nckap1l. All three of these homologs have the potential for phenotypes in the notochord. Below is a rough overview of our experimental plan.
Experimental plan
- Generate and validate mutants in G0 embryos.
- Phenotype notochord effects.
- Generate tissue-specific mutants. To evaluate whether the notochord effects are non-cell-autonomous, generate mutants using a notochord-specific promoter to express Cas9.
- Perform genetic rescue experiments using complementation with wild-type protein.
Methods
For details about the technical analyses described in this pub, see the associated pub describing our evaluation framework [1].
All code related to this pub is available in this GitHub repo (DOI: 10.5281/zenodo.15707938). Code specific to C. intestinalis is in these analysis notebooks and this set of visualization scripts.
Ciona identifier mapping
After consulting with experts, we learned that the Ciona community doesn’t use UniProt identifiers regularly in their work. For our organism selection work and in the Zoogle interface, we rely on these identifiers to distinguish proteins in the Ciona proteome. Thus, to bridge our data and relevant gene-expression resources, we needed maps between UniProt identifiers and other identifiers. The community uses multiple gene and protein identifiers for previous versions of the Ciona genome, including “KH” identifiers for the KH2012 genome (GCF_000224145.1) (used in resources like ANISEED [56]) and “KY” identifiers for the KY2021 genome [57] (used for a recently re-analyzed Ciona single-cell transcriptome, described at the top of the next section). To generate a map between the KY identifiers and UniProt identifiers, we ran all-versus-all BLASTp [58] using default parameters, setting the KY proteome as the database and the UniProt proteome as the query. For each KY protein, we selected the top UniProt identifier as a crude best match. We also obtained an Excel spreadsheet from Alberto Stolfi containing previously generated maps between the KY and KH identifiers, which we provide in our GitHub repo. Identifier mapping functionality can be found in our zoogletools Python package.
You can find a copy of the KH2012 to KY2021 identifier map here in our GitHub repository. The zoogletools Python package is available here.
Ciona single-cell developmental gene expression
We visualized single-cell RNA-seq data analyzed by Katarzyna Piekarz, as described in this GitHub repository using scripts in our zoogletools Python package. This dataset is a re-analysis of data from a 2019 paper by Cao et al. [59]. The major difference between the two datasets is that Cao et al. mapped their reads to the KH2012 genome, whereas Piekarz mapped their data to the KY2021 genome. In reusing the Piekarz data, we identified some discrepancies in file names, replicate labels, and developmental stage labels, which are corrected by helper scripts in the zoogletools package. You can find a description of those issues in this GitHub pull request. We’ve discussed some of these discrepancies with the authors of the Piekarz repo in this GitHub issue. There may be problems with stage labeling in the data released with the Cao et al. manuscript, with potential inconsistencies between the Gene Expression Omnibus deposition and the Broad Institute Single Cell Portal. We treated the cell labels in the portal dataset as a ground truth for the purposes of our exploration.
In addition to visualizing expression counts from the Piekarz re-analysis of the Cao et al. manuscript, we downloaded tissue type annotations from Cao et al. from the Broad Institute Single Cell Portal [60] and merged them into the Piekarz re-analysis. The zoogletools Python package also provides functionality for performing this merge operation.
Ciona bulk developmental gene expression
One major barrier to observing CRISPR knockout phenotypes can be the presence of high levels of maternally deposited RNA in Ciona embryos [61]. To understand the relative expression of genes across developmental stages, we visualized bulk RNA-seq data downloaded from the ANISEED database of Ciona gene expression [56]. This database uses the KH2012 genome for its identifiers. The zoogletools Python package provides scripts to generate gene expression visualizations from these data.
AI tool usage
We used Claude to help write code, clean up code, comment our code, draft text that we edited, suggest wording ideas for small phrases and sentence structures, and clarify and streamline text that we wrote. We also used Cursor to access Claude.
Visualization
We used plotly (v5.17.0) [62] arcadia-pycolor (v0.6.3) [63] to generate figures before manual adjustment.
Key takeaways
Advantages of Ciona as a disease model
During our diligence process, we came to realize a variety of advantages that Ciona has as a potential model for human diseases, including:
- Rapid development. Organoids derived from human iPSCs and mouse embryos have long differentiation or developmental timelines. Ciona embryos develop rapidly, reaching the larval stage overnight, allowing for rapid examination of genetic effects on diverse tissue systems during development.
- Morphogenetic processes. Ciona embryos undergo a variety of relevant morphogenetic processes, such as cell intercalation, division, lumen formation, and others. These allow scientists to probe the effects of genes and drugs on such processes.
- Ease of genetic experimentation. Ciona embryos can be subjected to transgenic overexpression, tissue-specific expression, RNAi, and gene knockout using electroporation or microinjection approaches. Moreover, Ciona embryos can be collected in the thousands, allowing for experimentation at scale.
Challenges of working with community resources
A major hurdle to our diligence work was learning how to access and use different resources maintained by the Ciona community. Resources such as the Ghost database [64] and ANISEED [56] contain a wealth of information, but use community-derived identifiers (see this explanation) that aren’t easy to understand without speaking with experts who can describe their history.
In addition to resolving gene identifiers, we also faced challenges in understanding and using publicly available data from the Ciona community. For example, two analyses of the same dataset generated by Cao et al., one by the original authors and another by a community member, had conflicting labels for developmental stages and replicates. We weren’t able to determine the source of this discrepancy, which could have arisen within the original dataset.
Next steps
We winnowed our list down to three genes that are promising to investigate in Ciona. Others who evaluate the same genes may arrive at different conclusions about what's practical to pursue, based on their expertise. Among the genes we decided not to study, we’d encourage others to try to generate mutants or knockdowns of NAXE to see whether there's any temperature-dependent effect on development, and whether supplementation with niacin derivatives could rescue mutant phenotypes. We also encourage others to check out our working diligence document and comment on whether you agree with our feasibility determinations.
We’re funding downstream work in the Alberto Stolfi lab at the Georgia Institute of Technology, who'll also share their results openly during the course of their experiments. Stay tuned for more information.
References
Conklin, E. G. "The organization and cell-lineage of the ascidian egg/by Edwin G." Conklin.[Academy of Natural Sciences] (1905).
Satoh, Noriyuki. "Developmental biology of ascidians." (No Title) (1994).
Plotly Technologies Inc. (2015). Collaborative data science. https://plot.ly
Share your thoughts!
Feel free to provide feedback by commenting in the box at the bottom of this page or by posting about this work on social media. Please make all feedback public so other readers can benefit from the discussion.