Bacteriophages — or “phages” for short — are viruses that infect bacteria. They are extremely under-studied relative to the great richness of biological novelty that they represent, are highly abundant in most ecosystems, and serve as excellent test cases for developing novel computational and experimental techniques.
Phages’ pervasive use of non-canonical nucleic acid chemistries is especially interesting to us. DNA is often represented as a simple four-letter alphabet. However, there are actually way more than four types of DNA nucleotides out there
Why? Diverse bacterial immune systems have evolved to recognize and destroy phage genomes. This puts extraordinary evolutionary pressure on phages to subvert these defenses. By changing the chemical nature of their DNA through genome modification or use of non-standard nucleotides, phages fortify their genome against bacterial attack
Our overarching goal is to discover new and exciting nucleic acid chemistries from bacteriophages in natural ecosystems.
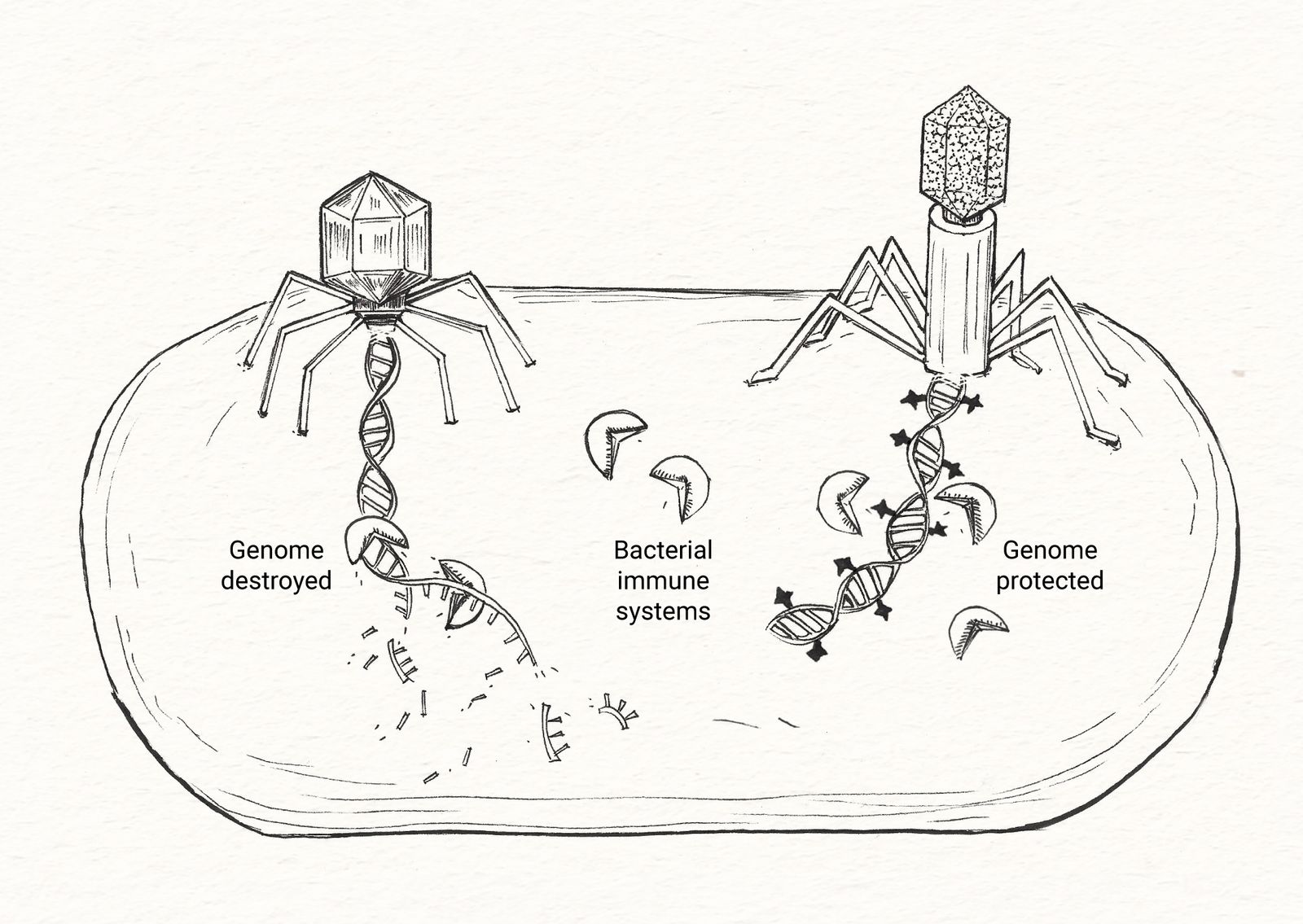
Phages protect their genomes with chemical modifications.
Bacteriophages are vulnerable to a wide variety of bacterial immune systems. Some immune systems work by chewing up the bacteriophage genome. In response, phages have evolved special, protective nucleic acid modifications that bacterial enzymes can't destroy. We suspect there is a rich diversity of nucleic acid chemistry waiting to be uncovered.
-
What omics tools can we leverage or adapt to learn more about the chemistry of phage nucleic acids in natural ecosystems?
-
How do we get more phages from the environment into the lab, where their nucleic acids can undergo detailed study?
-
And at the big-picture level, how can we bridge the gap between laboratory studies of isolated phages and omics-based studies of environmental phages?
We launched this project by getting our molecular biology and analytical techniques up and running on a couple laboratory-culturable phages with well-characterized DNA modifications. We’ve shared our methods for studying phage DNA, as well as the difficulties we encountered trying to adapt the methodology for phage RNA. We also worked on isolating phages from cheese microbial communities and screening them for genome modification using HPLC, and discovered one phage that uses a probable arabinose hypermodification of hydroxymethylcytosine. We experienced significant technical challenges adapting omics tools to screen communities for interesting DNA chemistries in high throughput. Ultimately, we have decided to ramp this project down because the technologies available to us today are not mature enough to let us quickly discover novel chemistries for commercialization. See more on this decision at the end of this page under “Discontinuing this project.”
We used phage T4 and phage SPO1 as our model non-canonical phage genomes. Phage T4 infects E. coli, and has unusual chemistry at its cytosines, where each cytosine is modified with a glucosyl-methyl group
Our first short pub details phage amplification and concentration, high-molecular-weight DNA extraction, and HPLC and MS analysis of phage nucleosides. We later added a section at the end of this pub discussing some of the technical challenges we encountered trying to apply modification detection methods to whole communities instead of isolated phages.
A workflow to isolate phage DNA and identify nucleosides by HPLC and mass spectrometry
This pub details a process for phage amplification and concentration, DNA extraction, and HPLC and MS analysis of phage nucleosides. We optimized the approach with model phages known to use non-canonical nucleosides in their DNA, but plan to apply it for other phages.
When we tried applying a similar approach to isolating phage RNA, we hit a few stumbling blocks. We describe these challenges and offer a possible path forward here:
Challenges of isolating bacteriophage mRNA for chemical analysis
We struggled to isolate enough phage mRNA for HPLC as we searched for new nucleoside chemistries. Ribodepleting the abundant extracted rRNA introduced contaminating DNA, and we were still left with more bacterial mRNA than phage transcripts. We suggest an alternative approach.
Next, we moved away from studying model phages to trying to discover phages with novel genome chemistries from natural communities. To this end, we started building out a phage culture collection from cheese microbial communities. We analyzed phage genomes for unusual nucleoside chemistries by HPLC. We isolated 114 host bacterial strains, used them to isolate 17 phages, and out of the phages we were able to chemically analyze, only one had a non-standard nucleoside chemistry. This phage is T4-like, and likely uses hydroxymethylation and arabinosylation to modify its cytosines, a modification previously discovered in E. coli phage RB69
This isolation effort was a labor-intensive process with a proportionally small payoff, indicating we need some way to prioritize certain communities, phages, and/or hosts for isolation. Metagenomic data could theoretically help with that, but in looking at the metagenomic data for the community where this modified phage originated, we found that only few reads mapped to our modified phage. We are unsure if this reflects the actual abundance of the phage in the community or if it is a consequence of library prep processes being less efficient on non-canonical DNA chemistry. Either way, we concluded that community sequencing is not always a good indicator of phage culturing success and may not be an effective way to prioritize communities for isolation of phages with genome modifications.
Isolation of a phage with an arabinosylated genome from a cheese microbial community
We sampled cheese microbial communities to discover bacteriophages with unusual genome chemistries. We isolated 114 bacterial host strains and 17 phages, and identified one phage with a probable arabinose hypermodification of hydroxymethylcytosine.
We had a low success rate in discovering phages with modified genomes using host culture and phage isolation, and we aren’t confident that metagenomics can help guide isolation. For this project to succeed, we’d need other high-throughput methods to survey microbial communities with unusual nucleoside content.
We experimented with using LC-MS/MS to analyze the chemical content of DNA extracted from whole microbial communities and community viromes. However, we were met with technical challenges that make this infeasible
We think this science is really exciting and has lots of potential for discovery and tool development, and we’re excited about new approaches that other groups are developing. However, we don’t see an immediate path forward for Arcadia to continue studying phage DNA modifications. We need to be able to rapidly discover novel, commercially-actionable phage genome chemistries using the technologies available to us today. We’ve decided to ice this project due to technological limitations.